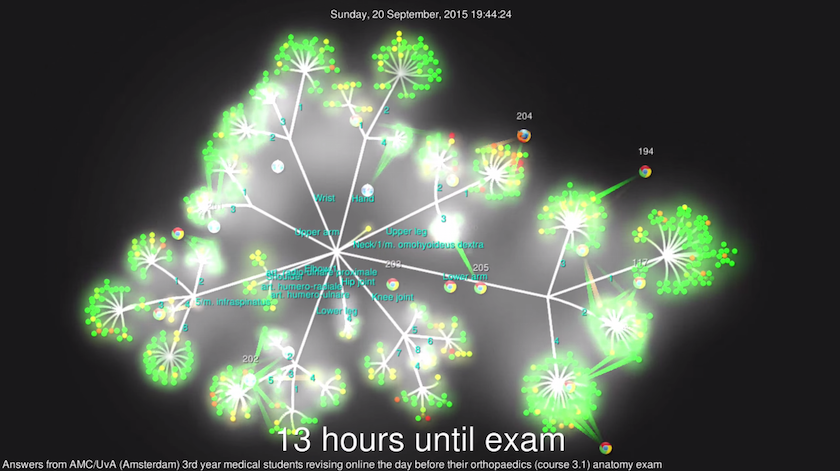
tl;dr: in the first year of medical school, I built an application that helps
fellow students and myself with studying anatomy. The answers of the last exam,
submitted by students while revising, have been visualized with Gource. Please
check out the YouTube video for the result:
Introduction to the application
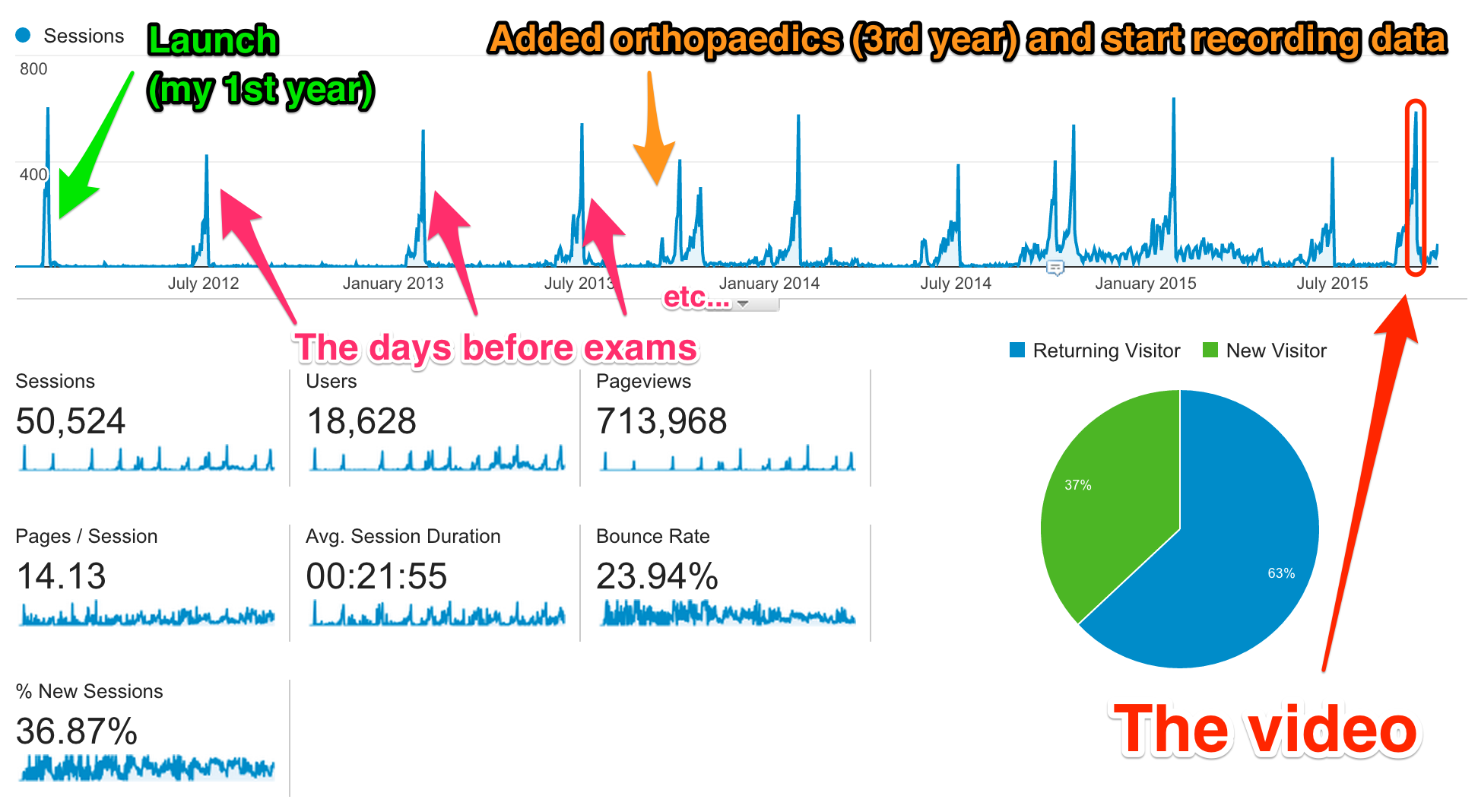
For medical students it is inevitable: you have to know all the anatomical terms by heart. The task is easy, but the amount of structures one has to learn is quite intimidating. I remember the feelings of despair that arose while staring at the latin words in the anatomy book. After one brave attempt, my attention span had decided: we needed a better way to study this (one that involves computers).
Building the visualization
This is the exam_date that we will be using for this visualization:
2015-09-21 08:30:00 +02:00Retrieving the answers
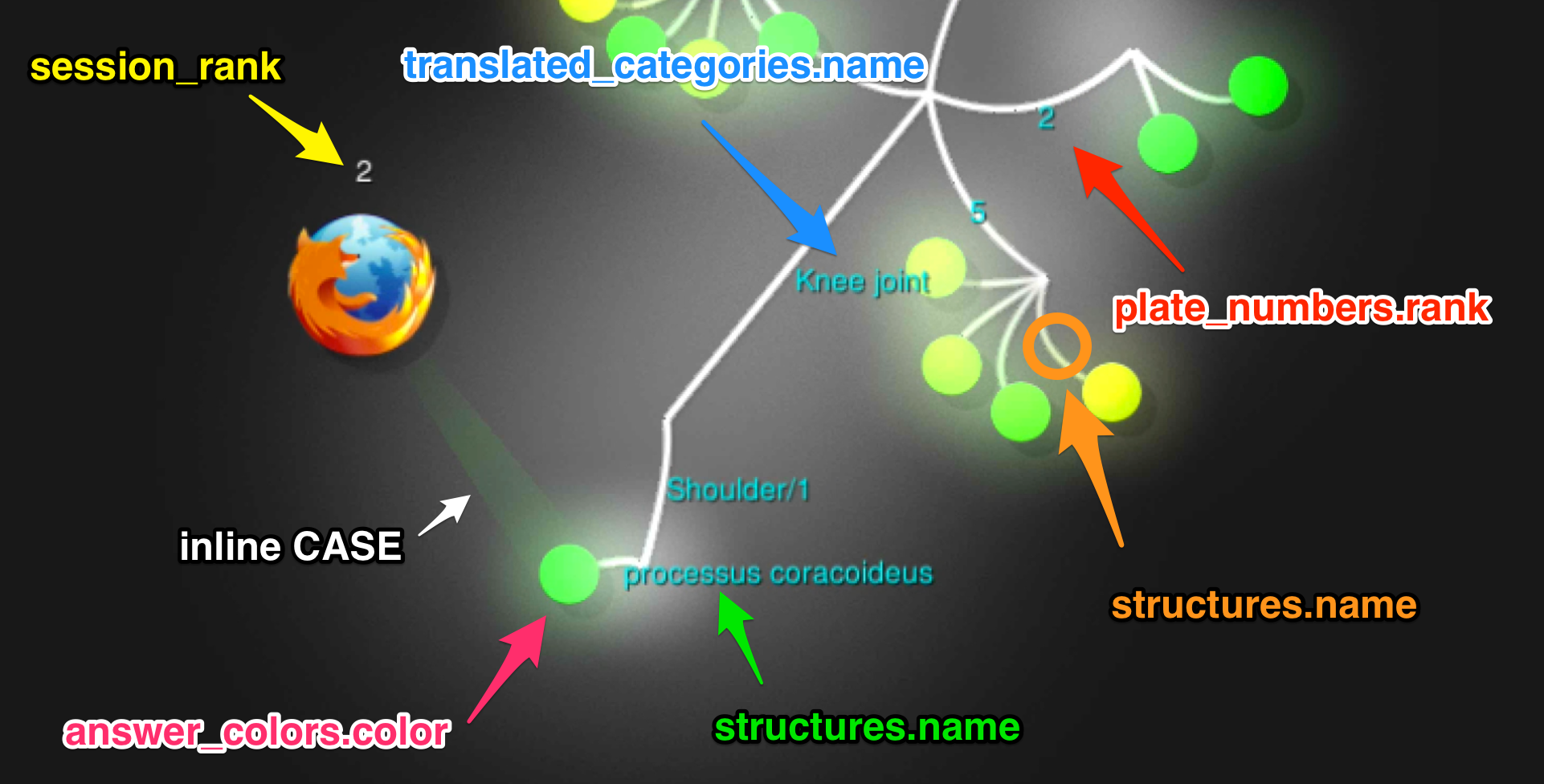
We use one big PostgreSQL query that yields all the answers from the timeframe in the right format. No scripting needed!
-- We use trigram similarity to determine answer correctness
CREATE EXTENSION IF NOT EXISTS pg_trgm;
WITH
translated_categories AS (
SELECT
id,
CASE name
WHEN 'Bovenarm' THEN 'Upper arm'
WHEN 'Bovenbeen' THEN 'Upper leg'
WHEN 'Elleboog' THEN 'Elbow'
WHEN 'Enkel' THEN 'Ankle'
WHEN 'Hals' THEN 'Neck'
WHEN 'Heupgewricht' THEN 'Hip joint'
WHEN 'Kniegewricht' THEN 'Knee joint'
WHEN 'Onderarm' THEN 'Lower arm'
WHEN 'Onderbeen' THEN 'Lower leg'
WHEN 'Pols' THEN 'Wrist'
WHEN 'Schouder' THEN 'Shoulder'
WHEN 'Voet' THEN 'Foot'
ELSE name
END AS name
FROM categories
),
answer_colors AS (
SELECT
step::float / 10 AS similarity,
CASE step
WHEN 10 THEN '00FF00' -- Green: 100% correct answer
WHEN 9 THEN '32FF00'
WHEN 8 THEN '65FF00'
WHEN 7 THEN '99FF00'
WHEN 6 THEN 'CCFF00'
WHEN 5 THEN 'FFFF00' -- Yellow: meh
WHEN 4 THEN 'FFCC00'
WHEN 3 THEN 'FF9900'
WHEN 2 THEN 'FF6600'
WHEN 1 THEN 'FF3200'
WHEN 0 THEN 'FF0000' -- Red: wrong answer :-(
END AS color
FROM generate_series(10, 0, -1) step
),
ranked_sessions AS (
SELECT
session_id,
rank() OVER (ORDER BY min(created_at)) session_rank
FROM answers
WHERE session_id IS NOT NULL
AND created_at
BETWEEN timestamp :exam_date - interval '28 hours'
AND :exam_date
GROUP BY session_id
),
plate_numbers AS (
SELECT
id,
category_id,
rank() OVER (PARTITION BY category_id ORDER BY plates.id)
FROM plates
)
SELECT
round(extract(epoch from answers.created_at)),
session_rank,
-- Green 'beam' (A) when the answer is 100% correct
CASE similarity(answers.input, structures.name)
WHEN 1 THEN 'A'
ELSE 'M'
END,
concat_ws('/',
translated_categories.name,
plate_numbers.rank,
structures.name,
rank() OVER (PARTITION BY structure_id ORDER BY answers)
),
color
FROM answers
JOIN structures ON structures.id = structure_id
JOIN answer_colors
ON answer_colors.similarity =
round(similarity(input, structures.name)::numeric, 1)
JOIN plate_numbers ON plate_numbers.id = plate_id
JOIN ranked_sessions rs ON rs.session_id = answers.session_id
JOIN translated_categories ON translated_categories.id = category_id
WHERE answers.created_at
BETWEEN timestamp with time zone :exam_date - interval '28 hours'
AND timestamp with time zone :exam_date
ORDER BY answersThe results from the query seem to match Gource’s custom log format:
head -n 3 $ANSWERS_PATH
echo
tail -n 3 $ANSWERS_PATH1442729360|1|A|Knee joint/1/meniscus medialis/1|00FF00
1442729371|1|A|Knee joint/1/lig. cruciatum posterior/1|00FF00
1442729377|1|A|Knee joint/1/meniscus lateralis/1|00FF00
1442815272|271|M|Upper leg/4/m. biceps femoris caput longum/75|32FF00
1442815283|271|A|Upper leg/4/m. vastus lateralis/75|00FF00
1442815322|271|M|Upper leg/4/m. peroneus longus/71|FF3200How many answers do we have in total?
wc -l < $ANSWERS_PATH50687Captions
tail -n 3 $CAPTIONS_PATH1442809800|2 hours until exam
1442813400|1 hour until exam
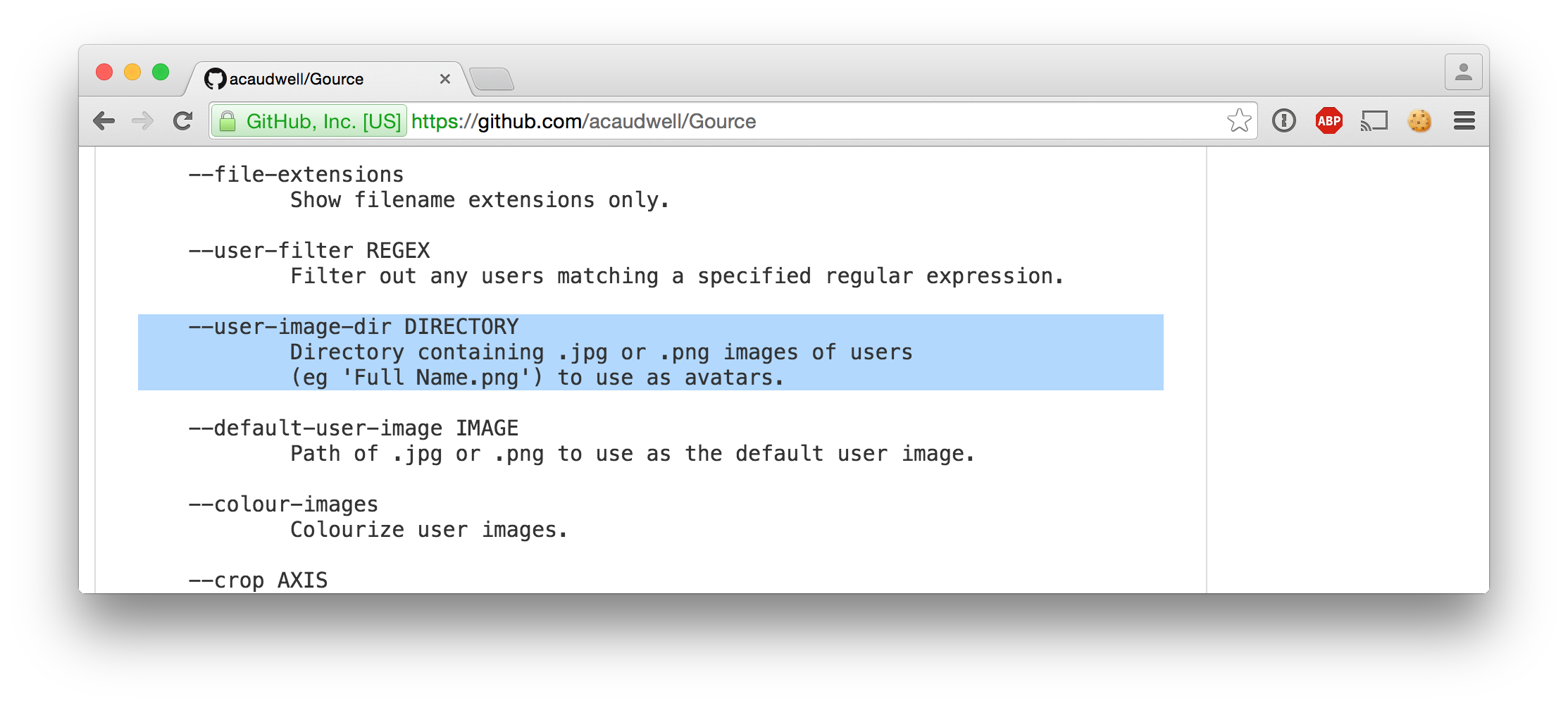
1442817000|Exam begins at 08:30...User images
Retrieving user agent data per session (rank)
SELECT
rank() OVER (ORDER BY min(created_at)) session_rank,
user_agent,
min(id) first_id,
min(created_at) session_start
FROM answers
WHERE session_id IS NOT NULL
AND answers.created_at
BETWEEN timestamp with time zone :exam_date - interval '28 hours'
AND timestamp with time zone :exam_date
GROUP BY session_id, user_agent1|Mozilla/5.0 (Macintosh; Intel Mac OS X 10_10_4) AppleWebKit/600.7.12 (KHTML, like Gecko) Version/8.0.7 Safari/600.7.12|1465360|2015-09-20 06:09:19.603637
2|Mozilla/5.0 (Windows NT 10.0; WOW64; rv:40.0) Gecko/20100101 Firefox/40.0|1465384|2015-09-20 06:19:55.221907
3|Mozilla/5.0 (Macintosh; Intel Mac OS X 10_10_5) AppleWebKit/600.8.9 (KHTML, like Gecko) Version/8.0.8 Safari/600.8.9|1465408|2015-09-20 06:28:14.890441Linking the sessions to browser icons
ls -l $USER_IMAGES_PATH/{1,2,3}.png | cut -d/ -f4-1.png -> /Users/pepijn/Desktop/browser_icons/Safari.png
2.png -> /Users/pepijn/Desktop/browser_icons/Firefox.png
3.png -> /Users/pepijn/Desktop/browser_icons/Safari.pngPutting it all together
time (gource -1280x720 \
--bloom-intensity 0.7 \
--caption-duration 15 \
--caption-file $CAPTIONS_PATH \
--caption-size 50 \
--dir-colour 00FFFF \
--dir-name-depth 2 \
--file-idle-time 10 \
--hide filenames \
--highlight-dirs \
--max-file-lag -1 \
--seconds-per-day 10000 \
--stop-at-end \
--title 'Answers from AMC/UvA (Amsterdam) 3rd year medical students revising online the day before their orthopaedics (course 3.1) anatomy exam' \
--user-image-dir $USER_IMAGES_PATH \
$ANSWERS_PATH 2>/dev/null) \
2>&1